Minimal Information Required In the Annotation of Models
![]() The Minimal Information Required In the Annotation of Models, initiated by the BioModels.net effort, aimed to produce a set of guidelines for the consistent annotation and curation of computational models in biology. It is suitable for use with any structured format for computational models.
The Minimal Information Required In the Annotation of Models, initiated by the BioModels.net effort, aimed to produce a set of guidelines for the consistent annotation and curation of computational models in biology. It is suitable for use with any structured format for computational models.
MIRIAM is a registered project of MIBBI (Minimum Information for Biological and Biomedical Investigations).
MIRIAM guidelines
MIRIAM guidelines are composed of three parts: #Reference correspondence, #Attribution annotation, and #External resource annotation. These are described below:
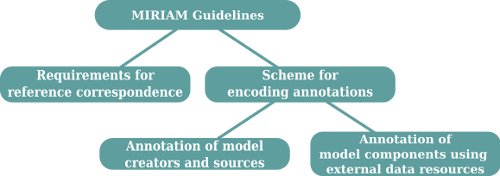
Reference correspondence
- The model must be encoded in a public, standardized, machine-readable format (SBML, CellML, GENESIS, ...).
- The model must comply with the standard in which it is encoded.
- The model must be clearly related to a single reference description. If a model is composed of different parts, it should still be accompanied with a description of the derived/combined model.
- The encoded model structure must reflect the biological processes described by the reference description.
- The model must be instantiable in a simulation: all quantitative attributes must be defined, including initial conditions.
- When instantiated, the model must be able to reproduce all results given in the reference description within an epsilon (algorithms, round-up errors).
Attribution annotation
- The model has to be named.
- A citation to the reference description must be provided (complete citation, unique identifier, unambiguous URL). The citation should identify the authors of the model.
- The name and contact information for model creators must be provided.
- The date and time of model creation and last modification should be specified. A history is useful but not required.
- The model should be linked to a precise statement about the terms of it's distribution. MIRIAM does not require “freedom of use” or “no cost”.
External resource annotation
- The annotation must unambiguously relate a piece of knowledge to a model constituent.
- The referenced information should be described using a triplet {collection, identifier, qualifier}:
- The annotation should be written as a Uniform Resource Identifier (URI).
- The identifier should be considered within the context of the framework of the collection.
- Collection namespace and identifier are be combined into a single URI, such as: http://identifiers.org/collection/identifier. For example: http://identifiers.org/uniprot/P62158.
- Qualifiers (optional) should refine the link between the model constituent and the piece of knowledge: “has a”, “is version of”, “is homolog to”, etc.
- The standard set of valid URIs is agreed upon by the community. A database and the associated API (Web Services) have been developed at the EBI to provide the generation and interpretation of URIs.
MIRIAM URIs
An important part of the MIRIAM guidelines consists of the controlled annotation of model components, based on Uniform Resource Identifiers (URIs). In order to support this task, a set of controlled URIs were created: MIRIAM URIs. These allowed the unique and unambiguous identification of a model component, in a stable and perennial manner. The #MIRIAM Registry and Identifiers.org system are a set of services and resources that provide support for generating, interpreting and resolving MIRIAM URIs.
MIRIAM URIs are composed of two main parts: the first defines a namespace that particular 'entities of the same type' may occupy. This is called a collection. The second component precisely identifies a given entity within this collection, called a record. For example, 'http://identifiers.org/pubmed/16333295' is the MIRIAM URI that identifies the publication of the MIRIAM Standard within the PubMed data collection. Here, 'http://identifiers.org/pubmed' defines the collection (PubMed), and '16333295' precisely identifies the record within it.
MIRIAM Registry and Identifiers.org
In order to enable the interoperability of this annotation scheme, the community has to agree upon a set of recognised collections. The MIRIAM Registry is an online service created to catalogue these collections, their URIs and the corresponding physical URLs or resources, whether they are controlled vocabularies or databases.
By using the MIRIAM Registry, one can (via Web Services) generate MIRIAM URIs (URN form), as well as resolve them (transform them into physical locations of the corresponding pieces of knowledge). Directly resolvable URIs, are also made available through Identifiers.org, which acts as a resolving layer above the the Registry. Both forms will be supported equally, with services provided to allow interconversion between them.
Publications
- Juty N., Le Novère N., Laibe C. (2012)
- Identifiers.org and MIRIAM Registry: community resources to provide persistent identification.
- Nucleic Acids Research, 40: D580-D586
- PubMed - Open Access
- Laibe C., Le Novère N. (2007)
- MIRIAM Resources: tools to generate and resolve robust cross-references in Systems Biology.
- BMC Systems Biology, 1: 58
- PubMed - Open Access
- Le Novère N, Courtot M, Laibe C (2007)
- Adding semantics in kinetics models of biochemical pathways.
- Proceedings of the 2nd International Symposium on experimental standard conditions of enzyme characterizations, available at http://www.beilstein-institut.de/index.php?id=196
- Le Novère N., Finney A., Hucka M., Bhalla U., Campagne F., Collado-Vides J., Crampin E., Halstead M., Klipp E., Mendes P., Nielsen P., Sauro H., Shapiro B., Snoep J.L., Spence H.D., Wanner B.L. (2005)
- Minimum Information Requested In the Annotation of biochemical Models (MIRIAM)
- Nature Biotechnology, 23: 1509-1515.
- PubMed - DOI
